Tracking SARS-CoV-2 variants in Kurdistan Region of Iraq.
DOI:
https://doi.org/10.21271/ZJPAS.35.5.12Keywords:
¬SARS-COV2, Covid-19, Spike protein, RBS, Viral infectionAbstract
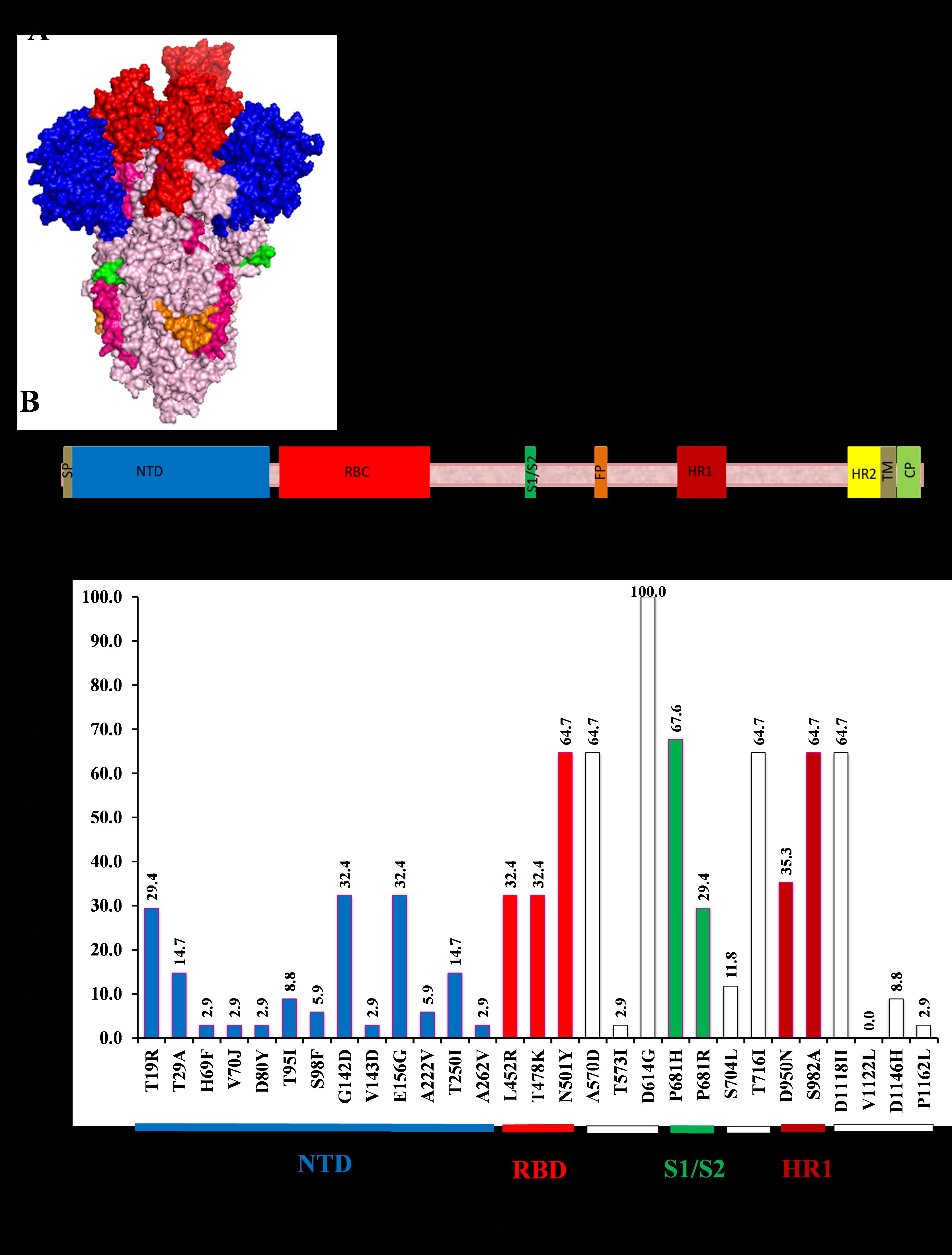
SARS COV-2, the etiology of the ongoing COVID-19 pandemic, is a positive single-stranded RNA virus which was first identified in the city of Wuhan in 2019. Since its first discovery, several variants of the virus have been reported around the world. Identifying the emerged SARS-CoV-2 variants overtime is important since this could alter the pathogenicity and transmissibility of the virus as well as the efficacy of the vaccine. Here, 34 genome sequence of SARS COV-2 samples, which were collected between Feb. 2021 to Dec. 2021 in Kurdistan Region of Iraq (KRG), were compared and analyzed in the spike (S) protein region. The nucleotide sequences were collected from the GISAID database (https://gisaid.org/), and the SARS-COV-2 variants were determined based on the mutations that they carry. Additionally, the frequency of each mutation was determined in the S protein region of the isolates. Overall, 29 mutations were detected in the sequence samples; the majority (44.8%) were located in the N-terminal domain (NTD) of the protein, whereas only 10.3% located in the ribosomal binding domain (RBD). D614G was found in all sequenced strains, followed by 67.6% mutation of P681H. Moreover, seven deletions were found in the NTD of the S protein, with 61.8% of the deletions was occurred at the position Y144. Alpha variants (B.1.1.7) were found to be dominant lineage during the period of this study, accounted for 65% of the sequence samples. Overall, our data support the speedy increase of the SARS-COV-2 genome diversity. Based on our knowledge, this is the first mega sequence analyses of different SARS-COV-2 strains from KRG of Iraq.
References
ABDULAH, D. M., QAZLI, S. S. A. & SULEMAN, S. K. 2021. Response of the public to preventive measures of COVID-19 in Iraqi Kurdistan. Disaster medicine and public health preparedness, 15, e17-e25.
ABDULLAH, H. M., ALI, S. M. & SABIR, D. K. 2020. Data Suggesting That COVID-19 May Have Existed in the Kurdistan Region of Iraq at the Time of the Outbreak in Wuhan Province of China. AJR Am J Roentgenol, 214, 1287-1294.
AHMED, J. Q., MAULUD, S. Q., AL-QADI, R., MOHAMED, T. A., TAYIB, G. A., HASSAN, A. M., TAHA, L. S., QASIM, K. M. & TAWFEEQ, M. A. 2022. Sequencing and mutations analysis of the first recorded SARS-CoV-2 Omicron variant during the fourth wave of pandemic in Iraq. Braz J Infect Dis, 102677.
ARORA, P., PÖHLMANN, S., HOFFMANN, M. J. S. T. & THERAPY, T. 2021. Mutation D614G increases SARS-CoV-2 transmission. 6, 1-2.
ASSESSMENT, R. R. J. E. C. F. D. P. & CONTROL 2020. Detection of new SARS-CoV-2 variants related to mink. European Centre for Disease Prevention and Control.
BANGARU, S., OZOROWSKI, G., TURNER, H. L., ANTANASIJEVIC, A., HUANG, D., WANG, X., TORRES, J. L., DIEDRICH, J. K., TIAN, J.-H. & PORTNOFF, A. D. J. S. 2020. Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate. 370, 1089-1094.
BRIEF, T. A. 2021. Emergence of SARS-CoV-2 B. 1.617 variants in India and situation in the EU/EEA. Frösunda, Sweden: European Centre for Disease Prevention and Control.
CAPOZZI, L., SIMONE, D., BIANCO, A., DEL SAMBRO, L., RONDINONE, V., PACE, L., MANZULLI, V., IACOBELLIS, M. & PARISI, A. 2021. Emerging Mutations Potentially Related to SARS-CoV-2 Immune Escape: The Case of a Long-Term Patient. Life (Basel), 11.
CHOI, J. Y. & SMITH, D. M. 2021. SARS-CoV-2 variants of concern. Yonsei medical journal, 62, 961.
CIOTTI, M., ANGELETTI, S., MINIERI, M., GIOVANNETTI, M., BENVENUTO, D., PASCARELLA, S., SAGNELLI, C., BIANCHI, M., BERNARDINI, S. & CICCOZZI, M. 2019. COVID-19 outbreak: an overview. Chemotherapy, 64, 215-223.
DAVIES, N. G., ABBOTT, S., BARNARD, R. C., JARVIS, C. I., KUCHARSKI, A. J., MUNDAY, J. D., PEARSON, C. A. B., RUSSELL, T. W., TULLY, D. C., WASHBURNE, A. D., WENSELEERS, T., GIMMA, A., WAITES, W., WONG, K. L. M., VAN ZANDVOORT, K., SILVERMAN, J. D., GROUP, C. C.-W., CONSORTIUM, C.-G. U., DIAZ-ORDAZ, K., KEOGH, R., EGGO, R. M., FUNK, S., JIT, M., ATKINS, K. E. & EDMUNDS, W. J. 2021. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science, 372.
DOERKSEN, T., LU, A., NOLL, L., ALMES, K., BAI, J., UPCHURCH, D. & PALINSKI, R. J. V. 2021. Near-Complete Genome of SARS-CoV-2 Delta (AY. 3) Variant Identified in a Dog in Kansas, USA. 13, 2104.
DUAN, L., ZHENG, Q., ZHANG, H., NIU, Y., LOU, Y. & WANG, H. 2020. The SARS-CoV-2 spike glycoprotein biosynthesis, structure, function, and antigenicity: implications for the design of spike-based vaccine immunogens. Frontiers in immunology, 11, 2593.
FARIA, N. R., MELLAN, T. A., WHITTAKER, C., CLARO, I. M., CANDIDO, D. D. S., MISHRA, S., CRISPIM, M. A., SALES, F. C., HAWRYLUK, I. & MCCRONE, J. T. 2021. Genomics and epidemiology of the P. 1 SARS-CoV-2 lineage in Manaus, Brazil. Science, 372, 815-821.
FRAMPTON, D., RAMPLING, T., CROSS, A., BAILEY, H., HEANEY, J., BYOTT, M., SCOTT, R., SCONZA, R., PRICE, J. & MARGARITIS, M. 2021. Genomic Characteristics and Clinical Impact of the Emergent SARS-CoV-2 B. 1.1. 7 Lineage in North Central London, November to December 2020.
GOWRISANKAR, A., PRIYANKA, T. & BANERJEE, S. 2022. Omicron: a mysterious variant of concern. The European Physical Journal Plus, 137, 1-8.
GURUPRASAD, K. 2020. Amino acid mutations in the protein sequences of human SARS CoV-2 Indian isolates compared to Wuhan-Hu-1 reference isolate from China.
HEMLALI, M., CHOUATI, T., GHAMMAZ, H., MELLOUL, M., ALAOUI AMINE, S., RHOULAM, S., TOUIL, N., ENNIBI, K., OUMZIL, H. & MOHAMED, R. J. M. R. A. 2022. Coding-Complete Genome Sequences of a Delta Subvariant (AY. 33) of SARS-CoV-2 Obtained from Moroccan COVID-19 Patients. 11, e01099-21.
HU, J., PENG, P., CAO, X., WU, K., CHEN, J., WANG, K., TANG, N. & HUANG, A.-L. 2022a. Increased immune escape of the new SARS-CoV-2 variant of concern Omicron. Cellular & Molecular Immunology, 19, 293-295.
HU, J., PENG, P., CAO, X., WU, K., CHEN, J., WANG, K., TANG, N., HUANG, A.-L. J. C. & IMMUNOLOGY, M. 2022b. Increased immune escape of the new SARS-CoV-2 variant of concern Omicron. 1-3.
JOHNSON, B. A., XIE, X., BAILEY, A. L., KALVERAM, B., LOKUGAMAGE, K. G., MURUATO, A., ZOU, J., ZHANG, X., JUELICH, T., SMITH, J. K., ZHANG, L., BOPP, N., SCHINDEWOLF, C., VU, M., VANDERHEIDEN, A., WINKLER, E. S., SWETNAM, D., PLANTE, J. A., AGUILAR, P., PLANTE, K. S., POPOV, V., LEE, B., WEAVER, S. C., SUTHAR, M. S., ROUTH, A. L., REN, P., KU, Z., AN, Z., DEBBINK, K., DIAMOND, M. S., SHI, P. Y., FREIBERG, A. N. & MENACHERY, V. D. 2021. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature, 591, 293-299.
KEMP, S. A., MENG, B., FERRIERA, I. A. T. M., DATIR, R., HARVEY, W. T., PAPA, G., LYTRAS, S., COLLIER, D. A., MOHAMED, A., GALLO, G., THAKUR, N., CARABELLI, A. M., KENYON, J. C., LEVER, A. M., DE MARCO, A., SALIBA, C., CULAP, K., CAMERONI, E., PICCOLI, L., CORTI, D., JAMES, L. C., BAILEY, D., ROBERTSON, D. L. & GUPTA, R. K. 2021. Recurrent emergence and transmission of a SARS-CoV-2 spike deletion H69/V70. BioRxiv
KHATEEB, J., LI, Y. & ZHANG, H. J. C. C. 2021. Emerging SARS-CoV-2 variants of concern and potential intervention approaches. 25, 1-8.
LAURING, A. S. & HODCROFT, E. B. 2021. Genetic variants of SARS-CoV-2—what do they mean? Jama, 325, 529-531.
LI, Q., WU, J., NIE, J., ZHANG, L., HAO, H., LIU, S., ZHAO, C., ZHANG, Q., LIU, H., NIE, L., QIN, H., WANG, M., LU, Q., LI, X., SUN, Q., LIU, J., ZHANG, L., LI, X., HUANG, W. & WANG, Y. 2020a. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell, 182, 1284-1294 e9.
LI, X., SONG, Y., WONG, G. & CUI, J. J. S. C. L. S. 2020b. Bat origin of a new human coronavirus: there and back again. 63, 461.
LIMA, A. R. J., RIBEIRO, G., VIALA, V. L., LIMA, L. P. O., MARTINS, A. J., BARROS, C., MARQUEZE, E., BERNARDINO, J., MORETTI, D. & RODRIGUES, E. S. J. M. 2021. SARS-CoV-2 genomic monitoring in the São Paulo State unveils new sublineages of the AY. 43 strain.
MANSBACH, R. A., CHAKRABORTY, S., NGUYEN, K., MONTEFIORI, D. C., KORBER, B. & GNANAKARAN, S. 2021. The SARS-CoV-2 Spike variant D614G favors an open conformational state. Science advances, 7, eabf3671.
MCCARTHY, K. R., RENNICK, L. J., NAMBULLI, S., ROBINSON-MCCARTHY, L. R., BAIN, W. G., HAIDAR, G. & DUPREX, W. P. J. S. 2021. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science, 371, 1139-1142.
MENG, B., KEMP, S. A., PAPA, G., DATIR, R., FERREIRA, I., MARELLI, S., HARVEY, W. T., LYTRAS, S., MOHAMED, A., GALLO, G., THAKUR, N., COLLIER, D. A., MLCOCHOVA, P., CONSORTIUM, C.-G. U., DUNCAN, L. M., CARABELLI, A. M., KENYON, J. C., LEVER, A. M., DE MARCO, A., SALIBA, C., CULAP, K., CAMERONI, E., MATHESON, N. J., PICCOLI, L., CORTI, D., JAMES, L. C., ROBERTSON, D. L., BAILEY, D. & GUPTA, R. K. 2021. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep, 35, 109292.
MOHAMMAD, P., AMINE, S., SABIR, D., SAEED, F., HAMA AMIN, D. J. H. E. & PROMOTION, H. 2021. Psychological Impact of COVID-19 on Healthcare Workers in the Kurdistan Region, Iraq. Health Education and Health Promotion, 9, 303-308.
PLANAS, D., VEYER, D., BAIDALIUK, A., STAROPOLI, I., GUIVEL-BENHASSINE, F., RAJAH, M. M., PLANCHAIS, C., PORROT, F., ROBILLARD, N., PUECH, J., PROT, M., GALLAIS, F., GANTNER, P., VELAY, A., LE GUEN, J., KASSIS-CHIKHANI, N., EDRISS, D., BELEC, L., SEVE, A., COURTELLEMONT, L., PERE, H., HOCQUELOUX, L., FAFI-KREMER, S., PRAZUCK, T., MOUQUET, H., BRUEL, T., SIMON-LORIERE, E., REY, F. A. & SCHWARTZ, O. 2021a. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature, 596, 276-280.
PLANAS, D., VEYER, D., BAIDALIUK, A., STAROPOLI, I., GUIVEL-BENHASSINE, F., RAJAH, M. M., PLANCHAIS, C., PORROT, F., ROBILLARD, N. & PUECH, J. J. N. 2021b. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. 596, 276-280.
PLANTE, J. A., LIU, Y., LIU, J., XIA, H., JOHNSON, B. A., LOKUGAMAGE, K. G., ZHANG, X., MURUATO, A. E., ZOU, J. & FONTES-GARFIAS, C. R. 2021a. Spike mutation D614G alters SARS-CoV-2 fitness. Nature, 592, 116-121.
PLANTE, J. A., LIU, Y., LIU, J., XIA, H., JOHNSON, B. A., LOKUGAMAGE, K. G., ZHANG, X., MURUATO, A. E., ZOU, J. & FONTES-GARFIAS, C. R. J. N. 2021b. Spike mutation D614G alters SARS-CoV-2 fitness. 592, 116-121.
POUDEL, S., ISHAK, A., PEREZ-FERNANDEZ, J., GARCIA, E., LEÓN-FIGUEROA, D. A., ROMANÍ, L., BONILLA-ALDANA, D. K. & RODRIGUEZ-MORALES, A. J. 2022. Highly mutated SARS-CoV-2 Omicron variant sparks significant concern among global experts–What is known so far? Travel medicine and infectious disease, 45, 102234.
RAO, R., DASS, M., MATHAI, D. & KUMAR, M. Prof Dr MV.
RASHID, P. M. A. & SALIH, G. F. 2022. Molecular and computational analysis of spike protein of newly emerged omicron variant in comparison to the delta variant of SARS-CoV-2 in Iraq. Mol Biol Rep, 49, 7437-7445.
SABIR, D. K., KHWARAHM, N. R., ALI, S. M., ABDOUL, H. J., MAHMOOD, K. I. & KODZIUS, R. J. J. O. I. S. 2020. Children protection against COVID-19 at the pandemic outbreak. Journal of Immunological Sciences 4.
SABIR, D. K. J. G. R. 2022. Analysis of SARS-COV2 spike protein variants among Iraqi isolates. 26, 101420.
SHANG, J., YE, G., SHI, K., WAN, Y., LUO, C., AIHARA, H., GENG, Q., AUERBACH, A. & LI, F. J. N. 2020. Structural basis of receptor recognition by SARS-CoV-2. 581, 221-224.
SHEN, X., TANG, H., PAJON, R., SMITH, G., GLENN, G. M., SHI, W., KORBER, B. & MONTEFIORI, D. C. J. N. E. J. O. M. 2021. Neutralization of SARS-CoV-2 variants B. 1.429 and B. 1.351. 384, 2352-2354.
SHU, Y. & MCCAULEY, J. 2017. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill, 22.
SIDIQ, K. 2021. Prevalence of COVID-19 and Possible Antigenic Drifts in SARS-CoV-2 Spike Protein in Kurdistan Region-Iraq. Passer Journal of Basic and Applied Sciences, 3, 0-0.
SINGH, P., SHARMA, K., SINGH, P., BHARGAVA, A., NEGI, S. S., SHARMA, P., BHISE, M., TRIPATHI, M. K., JINDAL, A. & NAGARKAR, N. M. 2022a. Genomic characterization unravelling the causative role of SARS-CoV-2 Delta variant of lineage B.1.617.2 in 2nd wave of COVID-19 pandemic in Chhattisgarh, India. Microb Pathog, 164, 105404.
SINGH, P., SHARMA, K., SINGH, P., BHARGAVA, A., NEGI, S. S., SHARMA, P., BHISE, M., TRIPATHI, M. K., JINDAL, A. & NAGARKAR, N. M. J. M. P. 2022b. Genomic characterization unravelling the causative role of SARS-CoV-2 Delta variant of lineage B. 1.617. 2 in 2nd wave of COVID-19 pandemic in Chhattisgarh, India. 105404.
SMALLMAN-RAYNOR, M. R. & CLIFF, A. D. J. M. 2022. Spatial Growth Rate of Emerging SARS-CoV-2 Lineages in England, September 2020-December 2021.
STIRRUP, O., BOSHIER, F., VENTURINI, C., GUERRA-ASSUNCAO, J. A., ALCOLEA-MEDINA, A., BECKETT, A., CHARALAMPOUS, T., DA SILVA FILIPE, A., GLAYSHER, S., KHAN, T., KULASEGARAN SHYLINI, R., KELE, B., MONAHAN, I., MOLLETT, G., PARKER, M., PELOSI, E., RANDELL, P., ROY, S., TAYLOR, J., WELLER, S., WILSON-DAVIES, E., WADE, P., WILLIAMS, R., CONSORTIUM, C.-U.-H. V. S., CONSORTIUM, C.-G. U., COPAS, A., CUTINO-MOGUEL, M. T., FREEMANTLE, N., HAYWARD, A. C., HOLMES, A., HUGHES, J., MAHUNGU, T., NEBBIA, G., PARTRIDGE, D., POPE, C., PRICE, J., ROBSON, S., SAEED, K., DE SILVA, T., SNELL, L., THOMSON, E., WITNEY, A. A. & BREUER, J. 2021. SARS-CoV-2 lineage B.1.1.7 is associated with greater disease severity among hospitalised women but not men: multicentre cohort study. BMJ Open Respir Res, 8.
SUPHANCHAIMAT, R., TEEKASAP, P., NITTAYASOOT, N., PHAIYAROM, M. & CETTHAKRIKUL, N. 2022. Forecasted trends of the new COVID-19 epidemic due to the Omicron variant in Thailand, 2022. bioRxiv.
TEGALLY, H., WILKINSON, E., GIOVANETTI, M., IRANZADEH, A., FONSECA, V., GIANDHARI, J., DOOLABH, D., PILLAY, S., SAN, E. J. & MSOMI, N. 2021. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature, 592, 438-443.
THOMSON, E. C., ROSEN, L. E., SHEPHERD, J. G., SPREAFICO, R., DA SILVA FILIPE, A., WOJCECHOWSKYJ, J. A., DAVIS, C., PICCOLI, L., PASCALL, D. J. & DILLEN, J. 2021. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell, 184, 1171-1187. e20.
UGUREL, O. M., MUTLU, O., SARIYER, E., KOCER, S., UGUREL, E., INCI, T. G., ATA, O. & TURGUT-BALIK, D. 2020. Evaluation of the potency of FDA-approved drugs on wild type and mutant SARS-CoV-2 helicase (Nsp13). Int J Biol Macromol, 163, 1687-1696.
VOLZ, E., HILL, V., MCCRONE, J. T., PRICE, A., JORGENSEN, D., O’TOOLE, Á., SOUTHGATE, J., JOHNSON, R., JACKSON, B. & NASCIMENTO, F. F. J. C. 2021. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. 184, 64-75. e11.
WANG, P., NAIR, M. S., LIU, L., IKETANI, S., LUO, Y., GUO, Y., WANG, M., YU, J., ZHANG, B. & KWONG, P. D. J. N. 2021. Antibody resistance of SARS-CoV-2 variants B. 1.351 and B. 1.1. 7. 593, 130-135.
WANG, R., HOZUMI, Y., YIN, C. & WEI, G.-W. 2020. Decoding SARS-CoV-2 transmission and evolution and ramifications for COVID-19 diagnosis, vaccine, and medicine. Journal of chemical information and modeling, 60, 5853-5865.
WISE, J. 2020. Covid-19: New coronavirus variant is identified in UK. British Medical Journal Publishing Group.
WU, F., ZHAO, S., YU, B., CHEN, Y.-M., WANG, W., SONG, Z.-G., HU, Y., TAO, Z.-W., TIAN, J.-H., PEI, Y.-Y., YUAN, M.-L., ZHANG, Y.-L., DAI, F.-H., LIU, Y., WANG, Q.-M., ZHENG, J.-J., XU, L., HOLMES, E. C. & ZHANG, Y.-Z. 2020. A new coronavirus associated with human respiratory disease in China. Nature, 579, 265-269.
XIA, X. 2021. Domains and Functions of Spike Protein in Sars-Cov-2 in the Context of Vaccine Design. Viruses, 13.
YURKOVETSKIY, L., WANG, X., PASCAL, K. E., TOMKINS-TINCH, C., NYALILE, T. P., WANG, Y., BAUM, A., DIEHL, W. E., DAUPHIN, A. & CARBONE, C. J. C. 2020. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. 183, 739-751. e8.
ZÁRATE, S., TABOADA, B., MUÑOZ-MEDINA, J. E., IŠA, P., SANCHEZ-FLORES, A., BOUKADIDA, C., HERRERA-ESTRELLA, A., SELEM MOJICA, N., ROSALES-RIVERA, M. & GÓMEZ-GIL, B. J. M. S. 2022. The Alpha variant (B. 1.1. 7) of SARS-CoV-2 failed to become dominant in Mexico. 10, e02240-21.
ZHOU, P., YANG, X.-L., WANG, X.-G., HU, B., ZHANG, L., ZHANG, W., SI, H.-R., ZHU, Y., LI, B. & HUANG, C.-L. 2020. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. bioRxiv.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2023 Pyman Mohamed Mohamedsalih, Karzan Jalal Salih, Banaz Omar Kareem, Dana Khdr Sabir

This work is licensed under a Creative Commons Attribution 4.0 International License.













