Automated Thalassemia cell image segmentation using hybrid Fuzzy C-Means and K-Means
DOI:
https://doi.org/10.21271/ZJPAS.35.4.03Keywords:
Thalassemia, color space conversion, segmentation, Fuzzy c-means, K-means, hybrid techniques.Abstract
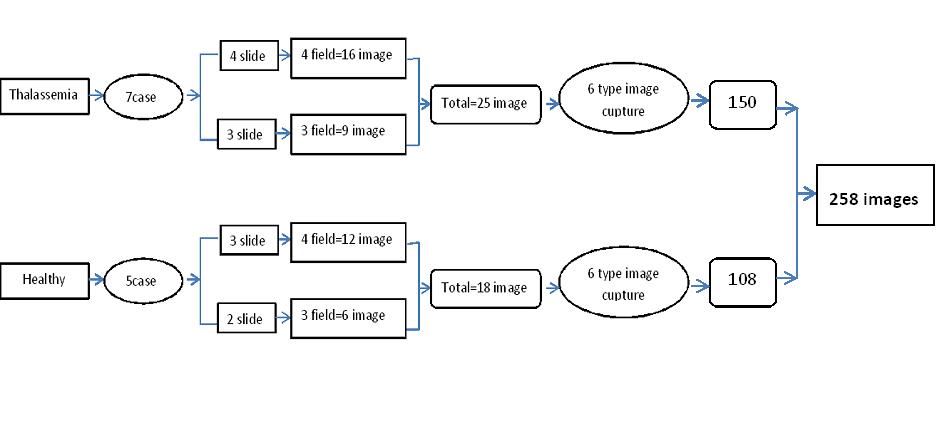
Thalassemia is a form of hereditary disease. Thalassemia is one of the world's most common illnesses. The morphology of red blood cells is most affected by this disorder. This research proposes a new method of automatically segmenting red blood cells from microscopic blood smear images. The research suggests a novel combination of image processing techniques and extensive preprocessing to achieve superior segmentation performance. In this work, the eleven designated color spaces, with six filters and three contrasts enhancing, Fuzzy c-means and K-means segmentation studied using five evaluation parameters. This evaluation is based on the ground truth image. The Photoshop program performs novel ground truth techniques for multi-object sense (RBC cells). The optimization of all image processing stages was obtained through local image datasets (258 images) obtained from seven thalassemia patients in the Erbil – thalassemia center and five samples of normal blood cells in Children Raparin Teaching Hospital. The image was captured with different light intensities (low, medium, high) and with /without a yellow filter in Biophysics Research lab /Education College / Salahaddin University –Erbil. This study found that the best light intensity for image slide capture utilizing a microscope was medium without using a yellow filter with an accuracy of 0.91± 0.14 and a performance of 95.34%.
References
Abood, Z. M., et al. (2017). Classification of red blood cells disease using fuzzy logic theory. 2017 International Conference on Current Research in Computer Science and Information Technology (ICCIT), IEEE.
Ahmad, I., et al. (2018). "Morphological features analysis for erythrocyte classification in IDA and thalassemia." International Journal of Advanced Computer Science and Applications 9(12).
Al-Dulaimi, K. A. K., et al. (2018). "Classification of white blood cell types from microscope images: Techniques and challenges." Microscopy science: Last approaches on educational programs and applied research (Microscopy Book Series, 8): 17-25.
Al-Hafiz, F., et al. (2018). "Red blood cell segmentation by thresholding and Canny detector." Procedia Computer Science 141: 327-334.
Çil, B., et al. (2020). "Discrimination of β-thalassemia and iron deficiency anemia through extreme learning machine and regularized extreme learning machine based decision support system." Medical Hypotheses 138: 109611.
Dybas, J., et al. (2022). "Trends in biomedical analysis of red blood cells–Raman spectroscopy against other spectroscopic, microscopic and classical techniques." TrAC Trends in Analytical Chemistry 146: 116481.
Gowda, S. N. and C. Yuan (2018). ColorNet: Investigating the importance of color spaces for image classification. Asian Conference on Computer Vision, Springer.
Hoshi, S., et al. (2022). "Improvement in Dacryoendoscopic Visibility after Image Processing Using Comb-Removal and Image-Sharpening Algorithms." Journal of clinical medicine 11(8): 2073.
Kumar, P. and K. S. Babulal (2022). "Hematological image analysis for segmentation and characterization of erythrocytes using FC-TriSDR." Multimedia Tools and Applications: 1-26.
Kumar, S. and D. Singh (2012). "Colorization of Gray Scale Images in YCbCr Color Space Using Texture Extraction and Luminance Mapping." IOSR Journal of Computer Engineering (IOSRJCE) 4(5): 27-32.
Lavanya, T. and S. Sushritha (2017). "Detection of sickle cell anemia and thalassemia using image processing techniques." International Journal of Innovative Science and Research Technology 2(12): 399-405.
Li, H., et al. (2020). "Color space transformation and multi-class weighted loss for adhesive white blood cell segmentation." IEEE access 8: 24808-24818.
Lin, Y.-H., et al. (2020). "Automatic detection and characterization of quantitative phase images of thalassemic red blood cells using a mask region-based convolutional neural network." Journal of Biomedical Optics 25(11): 116502.
Marzuki, N. I. B. C., et al. (2017). "Identification of thalassemia disorder using active contour." Indonesian Journal of Electrical Engineering and Computer Science 6(1): 160-165.
Matsuo, T., et al. (2013). Weighted Joint Bilateral Filter with Slope Depth Compensation Filter for Depth Map Refinement. VISAPP (2).
Mohamed, M. and B. Far (2012). An enhanced threshold based technique for white blood cells nuclei automatic segmentation. 2012 IEEE 14th International Conference on e-Health Networking, Applications and Services (Healthcom), IEEE.
Oussama, L., et al. (2017). "Red blood cell image enhancement techniques for cells with overlapping condition." Journal of Fundamental and Applied Sciences 9(4S): 614-628.
Patgiri, C. and A. Ganguly (2021). "Adaptive thresholding technique based classification of red blood cell and sickle cell using naíve bayes classifier and k-nearest neighbor classifier." Biomedical Signal Processing and Control 68: 102745.
Pellegrino, R. V., et al. (2021). Automated RBC Morphology Counting and Grading Using Image Processing and Support Vector Machine. 2021 IEEE 13th International Conference on Humanoid, Nanotechnology, Information Technology, Communication and Control, Environment, and Management (HNICEM), IEEE.
Prabha, D. S. and J. S. Kumar (2016). "Performance evaluation of image segmentation using objective methods." Indian J. Sci. Technol 9(8): 1-8.
Purwar, S., et al. (2021). Classification of thalassemia patients using a fusion of deep image and clinical features. 2021 11th International Conference on Cloud Computing, Data Science & Engineering (Confluence), IEEE.
Rashid, N. Z. N., et al. (2015). Unsupervised color image segmentation of red blood cell for thalassemia disease. 2015 2nd International Conference on Biomedical Engineering (ICoBE), IEEE.
Sadiq, S., et al. (2021). "Classification of β-Thalassemia Carriers From Red Blood Cell Indices Using Ensemble Classifier." IEEE access 9: 45528-45538.
Setsirichok, D., et al. (2012). "Classification of complete blood count and haemoglobin typing data by a C4. 5 decision tree, a naïve Bayes classifier and a multilayer perceptron for thalassaemia screening." Biomedical Signal Processing and Control 7(2): 202-212.
Sharif, J. M., et al. (2012). Red blood cell segmentation using masking and watershed algorithm: A preliminary study. 2012 international conference on biomedical engineering (ICoBE), IEEE.
Shirai, K., et al. (2022). "Adjoint Bilateral Filter and Its Application to Optimization-based Image Processing." APSIPA Transactions on Signal and Information Processing 11(1).
Tomari, R., et al. (2014). "Computer aided system for red blood cell classification in blood smear image." Procedia Computer Science 42: 206-213.
Tyas, D. A., et al. (2020). "Morphological, Texture, and Color Feature Analysis for Erythrocyte Classification in Thalassemia Cases." IEEE access 8: 69849-69860.
Tyas, D. A., et al. (2017). The classification of abnormal red blood cell on the minor thalassemia case using artificial neural network and convolutional neural network. Proceedings of the International Conference on Video and Image Processing.
Tyas, D. A., et al. (2022). "Erythrocyte (red blood cell) dataset in thalassemia case." Data in Brief 41: 107886.
Zou, K. H., et al. (2004). "Statistical validation of image segmentation quality based on a spatial overlap index1: scientific reports." Academic radiology 11(2): 178-189.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2023 Nabeel J. Ali, Assisst. Proff. Sardar Yaba

This work is licensed under a Creative Commons Attribution 4.0 International License.













